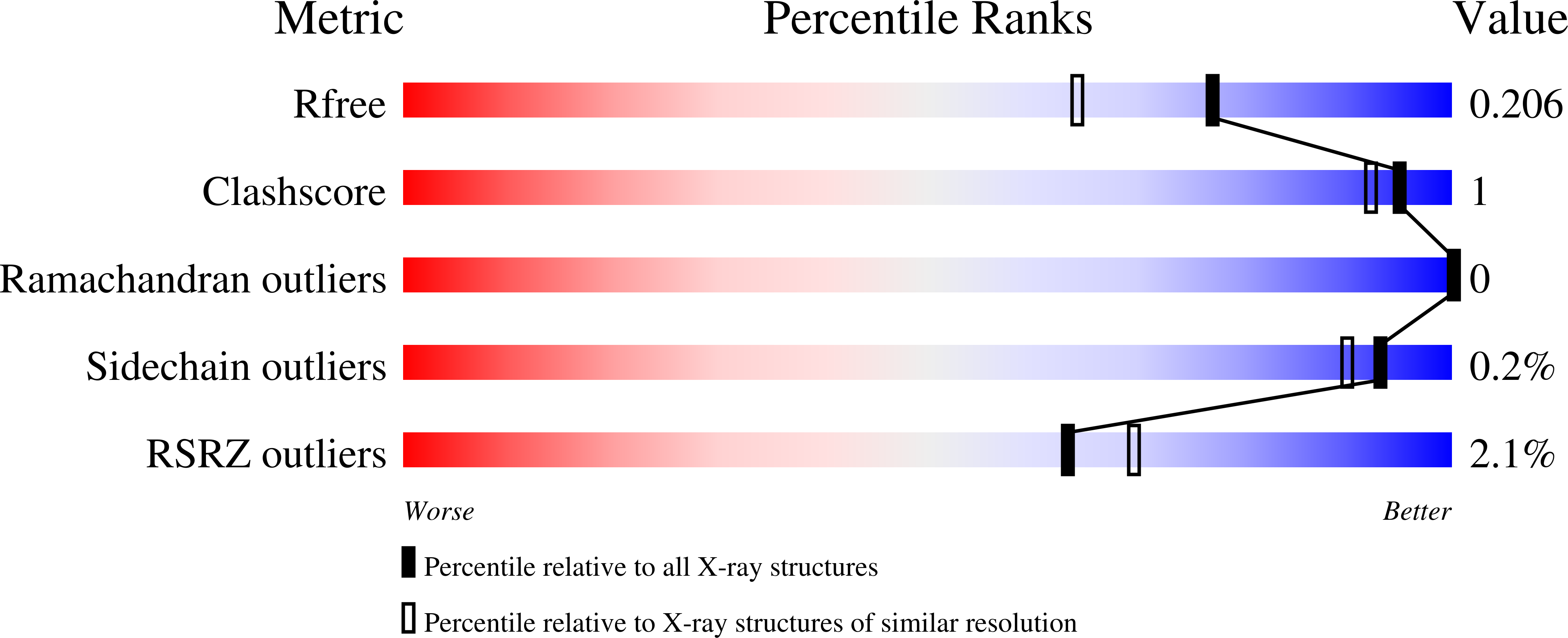
90P Query on 90P Download Ideal Coordinates CCD File D [auth A], N~2~-cyclohexyl-N~4~-(1-ethylpiperidin-4-yl)-6,7-dimethoxy-N~2~-methylquinazoline-2,4-diamine 24 H37 N5 O2 SAM Query on SAM Download Ideal Coordinates CCD File C [auth A], S-ADENOSYLMETHIONINE 15 H22 N6 O5 SDMS Query on DMS Download Ideal Coordinates CCD File Y [auth B] DIMETHYL SULFOXIDE 2 H6 O SZN Query on ZN Download Ideal Coordinates CCD File E [auth A]
E [auth A],
ZINC ION CL Query on CL Download Ideal Coordinates CCD File I [auth A], CHLORIDE ION UNX Query on UNX Download Ideal Coordinates CCD File AA [auth B]
AA [auth B],
UNKNOWN ATOM OR ION